We are active members of the California Institute for Quantitative Biosciences (QB3) and the Berkeley Stem Cell Center, with students in several departments including the UC Berkeley-UCSF Graduate Group in Bioengineering, Berkeley’s Biophysics Graduate Group, and Mechanical Engineering.

Node-Pore Sensing
Chang Chen, Andre Lai, Rachel Rex, Taylor Thomsen, Woochul Kim, Emilie Petit, William Rosas-Liu
Node-Pore Sensing (NPS) is a label-free technique we developed that can immunophenotype and mechanical phenotype single cells. We are advancing NPS such that immunophenotyping and mechanophenotyping can be performed within a single platform. Our collaborators include: Prof. Michael Lusting in EECS, with whom we are encoding our devices with codes used in radar, Wi-Fi, and telecommunications for real-time analysis and coincidence correction; Prof. Mark LaBarge from the City of Hope, with whom we are using mechano-NPS and machine learning to differentiate young vs. old human mammary epithelial cells (HMECs) and subsequently assess breast cancer risk; and Prof. Amanda Randles from Duke University, who is developing a "digital twin" of mechano-NPS to widen the scope of measured cell biophysical markers.
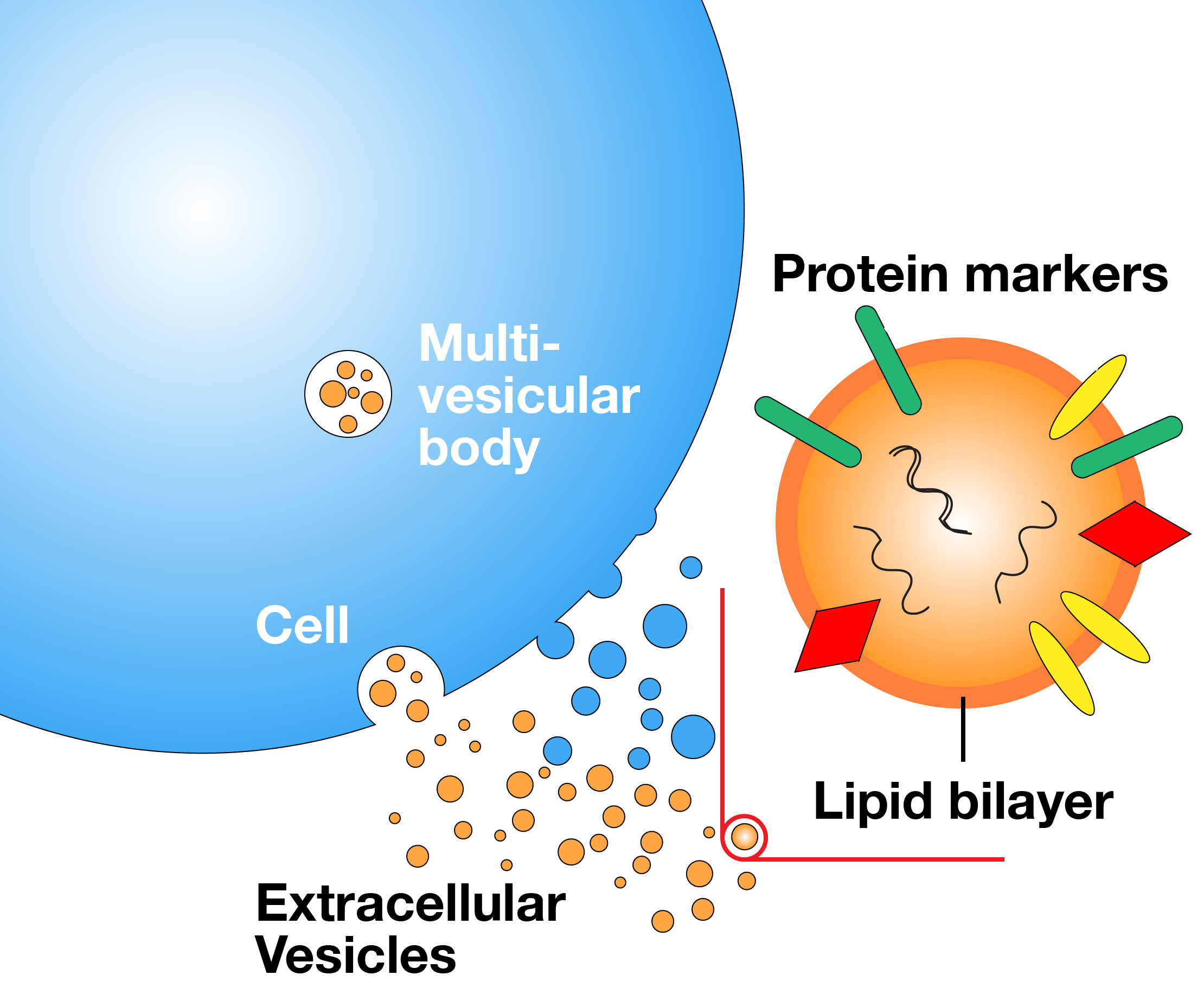
Extracellular Vesicle-Based Liquid Biopsy
Corey Pierce
We are developing a method to screen and quantify subpopulations of extracellular vesicles displaying tumor-specific protein markers. Our method could be used to monitor cancer patients and detect resistance to therapy.
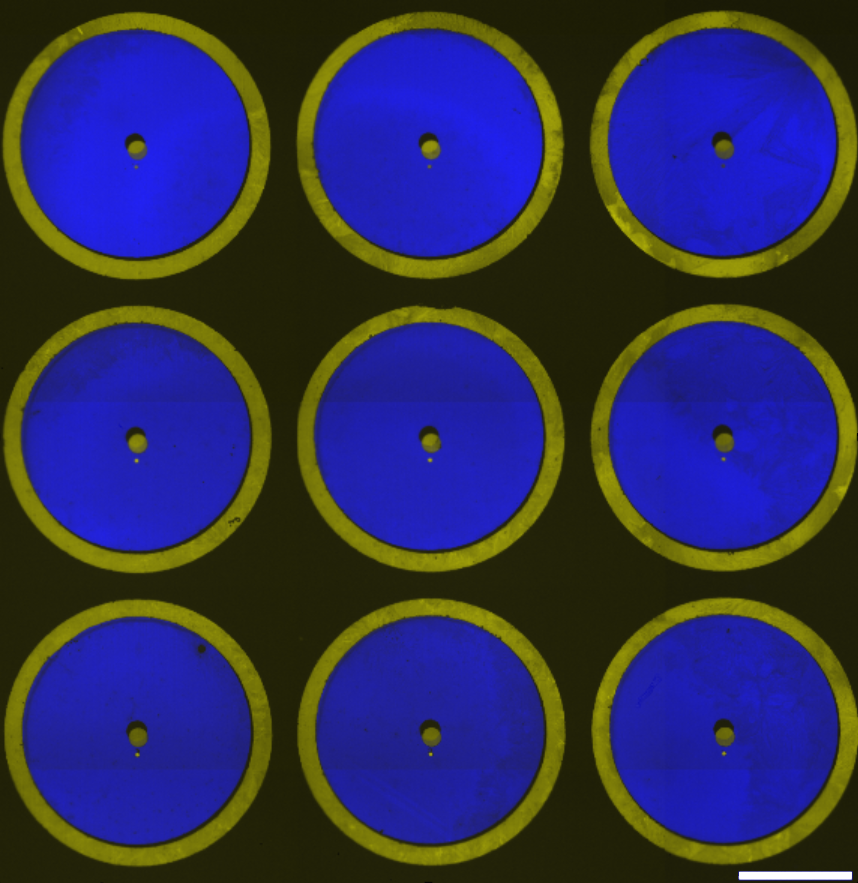
Studying the tumor microenvironment using high-throughput DNA-directed cell patterning
Pritika Acharya, Trinity Ellis, Maia Jeanneau,
DNA-directed cell patterning facilitates the high-resolution fabrication of different cellular systems in a high-throughput fashion. We are using this technique to study the contributions of different cell types to the tumor microenvironment. We are particularly interested in the influence of the immune cells in the breast tumor microenvironment, how different microenvironmental cells induce or support the epithelial-mesenychmal transition, and how microenvironmental factors govern the immunosuppressive mesenchymal stem cells.

Engineering Strategies for Studying the Neural Stem-Cell Niche
Olivia Scheideler
In a collaboration with Prof. David Schaffer (Chemical and Biomolecular Engineering), we are developing new DNA-based patterning technologies that enable us to dissect the complex mechanisms of niche signals on instructing single neural stem-cell fate decisions.
Patterning in Three Dimensions
We are developing methods to extend our high-throughput DNA patterning platform to three-dimensions.

Accelerated Aging in Cerebral Organoids
Stephanie Eberly, Virginia Yu
A critical limitation of using brain organoids to study aging mechanisms and to model neurodegenerative disease is that the neurons/tissue are immature, representing those of a fetal brain. To expand the reach of organoid technologies, we are developing cerebral organoids that undergo accelerating aging. Our success will enable researchers to investigate the real-time development and progression of age-associated neurodegenerative diseases such as Alzheimer’s, Parkinson’s, or Lou Gehrig’s disease.


